RESOURCES FOR PHYLOGENETICS WITH DNA BARCODES
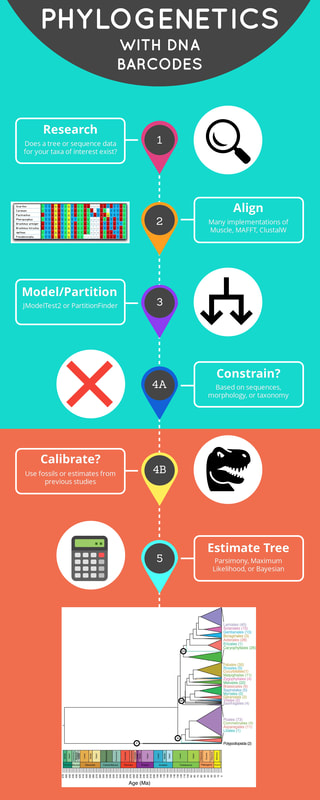
RESEARCH -Search literature to see if there are existing, useful phylogenies that you can just use. -Search sequence repositories for sequence data for taxa of interest: -BOLD (http://www.boldsystems.org) -Genbank (https://www.ncbi.nlm.nih.gov/genbank/) SEQUENCE ALIGNMENT -Align sequences to establish homology -Many implementations of Muscle, MAFFT, and ClustalW -Popular programs include Geneious, Mesquite, and R MODEL/PARTITION -Use model testing software to test different models of nucleotide substitution -To just do model testing use JModelTest2 (https://github.com/ddarriba/jmodeltest2) -Partition alignment by specifying different models for different genes or nucleotide positions -PartitionFinder2 can do model testing and partitioning simultaneously (http://www.robertlanfear.com/partitionfinder) CONSTRAIN TREE -Constraints can be specified to restrict the possible number of relationships among taxa that the phylogenetics software will explore -Constraining trees is generally a good idea for trees built from DNA barcodes, particularly if your taxon set includes taxa from divergent lineages (e.g. different families, orders, ect.) -Implementation of constraints depends on the program used to estimate the tree (read the manual) CALIBRATE TREE -There are two main ways of time calibrating a tree -Assign node ages using fossils and infer calibration during phylogenetic analysis -Rescale tree after phylogenetic analysis (Phylocom's Bladj) -Great tutorial on time calibration available from Tracy Heath (http://phyloworks.org/workshops/DivTime_BEAST2_tutorial_FBD.pdf) ESTIMATE TREE -Several different “flavors” of analysis (each with their own assumptions) including Parsimony, Maximum Likelihood, and Bayesian tree estimation -For Parsimony implementation use TNT (http://www.zmuc.dk/public/phylogeny/tnt/) -For Maximum Likelihood use RAxML(https://cme.h-its.org/exelixis/software.html) -For Bayesian use MrBayes (http://nbisweden.github.io/MrBayes/), BEAST (http://beast.community), BEAST2 (http://www.beast2.org) or RevBayes (https://revbayes.github.io) -Generally, people resist Parsimony at this point and prefer Maximum Likelihood or Bayesian tree estimation -You always have the option of using multiple approaches COMPUTING POWER -While many of these programs will run on local machines just fine for small sets of taxa, if you are doing analyses with hundreds of species or just want to do things faster, use CIPRES for free phylogenetics supercomputing (http://www.phylo.org)
1 Comment
Your comment will be posted after it is approved.
Leave a Reply. |
AuthorComputational resources kindly contributed and explained by members of our community. Archives
May 2023
Categories
All
|
 RSS Feed
RSS Feed
